Difference between revisions of "RhoA"
(New page: {{Subst:infobox}}) |
m |
||
| (10 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
{| class="toccolours" border="1" style="float: left; clear: right; margin: 0 0 1em 1em; border-collapse: collapse;" | {| class="toccolours" border="1" style="float: left; clear: right; margin: 0 0 1em 1em; border-collapse: collapse;" | ||
| − | ! | + | ! {infobox header}| '''{{PAGENAME}}''' |
|- | |- | ||
| − | | align="center" colspan="2" bgcolor="#ffffff" | [[Image: | + | | align="center" colspan="2" bgcolor="#ffffff" | [[Image:RhoA.JPG|200px|{{PAGENAME}}]] |
|- | |- | ||
| Substrate peptide name | | Substrate peptide name | ||
| Line 12: | Line 11: | ||
|- | |- | ||
| Determination type | | Determination type | ||
| − | | In situ | + | | In situ/In vitro |
|- | |- | ||
| Source | | Source | ||
| Line 21: | Line 20: | ||
|- | |- | ||
| Swissprot ID | | Swissprot ID | ||
| − | | [http://au.expasy.org/uniprot/ | + | | [http://au.expasy.org/uniprot/P61586 P61586 ] |
|- | |- | ||
| Reactive glutamines | | Reactive glutamines | ||
| − | | [[ | + | | [[TG2 reactive residues (RhoA)|Q52, Q63, Q136]] |
|- | |- | ||
| Reactive lysines | | Reactive lysines | ||
| − | | | + | | - |
|- | |- | ||
| Substrate sequence | | Substrate sequence | ||
| − | | | + | | EVDGK''Q''VELAL |
| + | |||
| + | WDTAG''Q''EDYDR | ||
| + | |||
| + | LAKMK''Q''EPVKP | ||
|- | |- | ||
| Structure | | Structure | ||
| − | | [http://www.rcsb.org/pdb/explore/explore.do?structureId= | + | | [http://www.rcsb.org/pdb/explore/explore.do?structureId=1A2B 1A2B] |
|- | |- | ||
| Surface accessibility | | Surface accessibility | ||
| − | | [http://gibk21.bse.kyutech.ac.jp/asaview/all/ | + | | [http://gibk21.bse.kyutech.ac.jp/asaview/all/1a2b.pdf ASAView] pdf |
| + | [http://gibk21.bse.kyutech.ac.jp/asaview/all/1a2b.txt ASAView] txt | ||
|- | |- | ||
| Disorder prediction | | Disorder prediction | ||
| Line 42: | Line 46: | ||
|- | |- | ||
| Reference | | Reference | ||
| − | | [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids= | + | | [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=9593707&dopt=Abstract PMID:9593707] |
|- | |- | ||
| Notes | | Notes | ||
| − | | | + | | Q63 is deamidated/transamidated by TG2 |
| + | |||
| + | The transamidated protein has increased binding affinity to ROCK-2. | ||
| + | |||
| + | The deamidated form (by cytotoxic dermonecrotizing factor or BTX) has decreased GTPase activity. | ||
| + | |- | ||
| + | |Video | ||
| + | |{{#ev:dailymotion|19dSThNNq2NLtkXZ8|300}} | ||
|- | |- | ||
| − | | | + | | {infobox header} | |
|- | |- | ||
|} | |} | ||
[[Category:Tissue transglutaminase|*]] | [[Category:Tissue transglutaminase|*]] | ||
| + | [[Category:Deamidase|*]] | ||
Latest revision as of 11:54, 10 March 2015
| RhoA | |
|---|---|
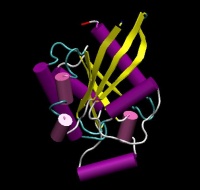
| |
| Substrate peptide name | RhoA |
| Synonyms | - |
| Determination type | In situ/In vitro |
| Source | Homo sapiens |
| Subcellular localization | Cytoskeleton |
| Swissprot ID | P61586 |
| Reactive glutamines | Q52, Q63, Q136 |
| Reactive lysines | - |
| Substrate sequence | EVDGKQVELAL
WDTAGQEDYDR LAKMKQEPVKP |
| Structure | 1A2B |
| Surface accessibility | ASAView pdf
ASAView txt |
| Disorder prediction | IUPred |
| Reference | PMID:9593707 |
| Notes | Q63 is deamidated/transamidated by TG2
The transamidated protein has increased binding affinity to ROCK-2. The deamidated form (by cytotoxic dermonecrotizing factor or BTX) has decreased GTPase activity. |
| Video | 19dSThNNq2NLtkXZ8|300}} |